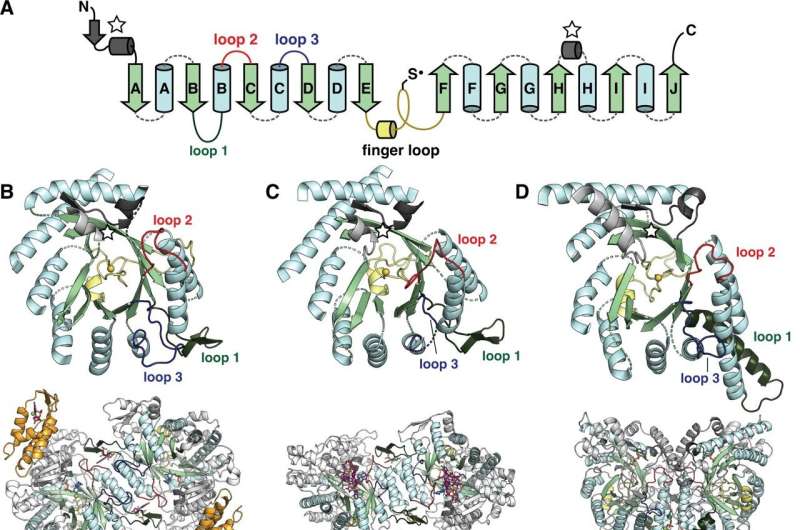
 The catalytic fold of the ribonucleotide reductase (RNR) family is a single 10-strand ɑ/β barrel, consisting of 10 β strands (light green) and 8 ɑ helices (light blue). (A) Each half of the cylinder contains a five-strand parallel β-sheet (βA-βE and βF-βJ) arranged in antiparallel orientation to each other. The two halves are joined by the so-called ‘finger loop’ (yellow) which usually begins with a short ɑ helix and contains a conserved cysteine which has been shown to be the site of the catalytically essential thiyl radical in all RNRs biochemically characterized. Diversity among RNRs is generated by N- and C-terminal extensions as well as insertions (dashed curves) between secondary structure elements in the ɑ/β barrel. Loops 1-3 (dark green, red, blue) have special names in the RNR literature for their involvement in regulating specificity. The gray secondary structure elements (stars) are partially embedded in the ɑ/β barrel and are involved in substrate binding. (B) The cylindrical part (top) of the Escherichia coli Class Ia catalytic subunit (lower dimer). The ATP cone domain is colored orange. (VS) The cylindrical part (top) of the Maritime Thermotoga Class II catalytic subunit (lower dimer). (D) The cylindrical part (top) of the class III catalytic subunit of phage T4 (dimer at the bottom). In class III RNRs, the loop 1 region (dark green) is a long helix involved in dimerization. Credit: eLife (2022). DOI: 10.7554/eLife.79790
The catalytic fold of the ribonucleotide reductase (RNR) family is a single 10-strand ɑ/β barrel, consisting of 10 β strands (light green) and 8 ɑ helices (light blue). (A) Each half of the cylinder contains a five-strand parallel β-sheet (βA-βE and βF-βJ) arranged in antiparallel orientation to each other. The two halves are joined by the so-called ‘finger loop’ (yellow) which usually begins with a short ɑ helix and contains a conserved cysteine which has been shown to be the site of the catalytically essential thiyl radical in all RNRs biochemically characterized. Diversity among RNRs is generated by N- and C-terminal extensions as well as insertions (dashed curves) between secondary structure elements in the ɑ/β barrel. Loops 1-3 (dark green, red, blue) have special names in the RNR literature for their involvement in regulating specificity. The gray secondary structure elements (stars) are partially embedded in the ɑ/β barrel and are involved in substrate binding. (B) The cylindrical part (top) of the Escherichia coli Class Ia catalytic subunit (lower dimer). The ATP cone domain is colored orange. (VS) The cylindrical part (top) of the Maritime Thermotoga Class II catalytic subunit (lower dimer). (D) The cylindrical part (top) of the class III catalytic subunit of phage T4 (dimer at the bottom). In class III RNRs, the loop 1 region (dark green) is a long helix involved in dimerization. Credit: eLife (2022). DOI: 10.7554/eLife.79790Cornell scientists have created an evolutionary model that links organisms living in today’s oxygen-rich atmosphere to a time billions of years ago when Earth’s atmosphere was low in oxygen – by analyzing ribonucleotide reductases (RNR), a family of proteins used by all free-living organisms and many viruses to repair and replicate DNA.
“By understanding the evolution of these proteins, we can understand how nature adapts to environmental changes at the molecular level. In return, we also learn about our planet’s past,” said Associate Professor Nozomi Ando. of Chemistry and Chemical Biology at the College of Arts and Sciences and corresponding author of the study. “Comprehensive phylogenetic analysis of ribonucleotide reductase family reveals ancestral clade” published in eLife October 4.
The co-first authors of the study are Audrey Burnim and Da Xu, PhD students in chemistry and chemical biology, and Matthew Spence, Research School of Chemistry, Australian National University, Canberra. Colin J. Jackson, Professor of Chemistry, Australian National University, Canberra, is a corresponding author.
This endeavor involved a large dataset of 6,779 RNR sequences; the phylogeny took several high-performance computers a combined seven months (1.4 million CPU hours) to calculate. Made possible by advances in computing, the approach opens up a new avenue for studying other diverse protein families that have evolutionary or medical significance.
RNRs have adapted to changes in the environment over billions of years to retain their catalytic mechanism due to their essential role for all DNA-based life, Ando said. His lab studies protein allostery, that is, how proteins are able to change activity in response to the environment. The evolutionary information in a phylogeny gives us a way to study the relationship between a protein’s primary sequence and its three-dimensional structure, dynamics, and function.
RNRs are thought to have ancient origins because they catalyze the reaction of converting RNA building blocks into DNA building blocks, Ando said, making them ideal for finding a molecular record.
“This chemistry would have been necessary to move from the hypothetical RNA world to the DNA/protein world we currently live in,” Ando said. “Based on the cofactors used by RNRs, it is also clear that this family of enzymes has adapted to the increase in oxygen in the Earth’s atmosphere. Both of these transitions took place billions ago. years.”
When scientists construct a phylogeny of a protein family, they calculate how currently existing sequences arose, Ando said. In this process, they have to estimate what happened in the past to get the sequences that exist now.
The researchers calculated the RNR phylogeny by collating a dataset of more than 100,000 sequences and narrowing down to a computable dataset of 6,779 sequences while maintaining the diversity of the entire family, Burnim said. The sequences vary in length from about 400 to 1100 amino acids. Using amino acid mutation models, they compared sequences to each other to determine when they diverged.
From this work, the researchers discovered a distinct new group of RNRs that would explain how two different adaptations to oxygen on Earth emerged within this family of proteins.
They used small-angle X-ray scattering at the Cornell High Energy Synchrotron Source, cryogenic electron microscopy at the Cornell Center for Materials Research, and the artificial intelligence program AlphaFold2 to study the RNR of the phage Synechococcus S-CBP4, a virus which infects a cyanobacterium, Xu said.
“When we calculated the family tree of RNRs, it turned out that there was a branch of RNRs that we didn’t know was a separate lineage,” Ando said. “This branch included sequences of marine organisms, including cyanophages. Our characterization of one of the sequences suggests that there was an early adaptation to oxygen. The connection of cyanophages was interesting because it suggests that their hosts (cyanobacteria) were present at the same time, and cyanobacteria are known to oxygenate the Earth.”
The findings support the idea that molecular adaptations to oxygen occurred much earlier than the planet’s large-scale environmental changes, as dated by geochemical records, Ando said.
This first-ever unified evolutionary model for all RNR classes could offer many future directions for the field, Xu said.
Ando plans to use the same approach to study how enzymes with the same overall structure have evolved to catalyze entirely different chemical reactions.
RNR ‘switch’ offers hope in fight against antibiotic resistant bacteria
Audrey A Burnim et al, A comprehensive phylogenetic analysis of the ribonucleotide reductase family reveals an ancestral clade, eLife (2022). DOI: 10.7554/eLife.79790
eLife
Provided by Cornell University
Quote: Protein family shows how life adapted to oxygen (October 4, 2022) Retrieved October 4, 2022 from https://phys.org/news/2022-10-protein-family-life-oxygen. html
This document is subject to copyright. Except for fair use for purposes of private study or research, no part may be reproduced without written permission. The content is provided for information only.
#Protein #family #shows #life #adapted #oxygen